The Why and How of Genetic Diversity

Whilst cichlid fish might look incredibly diverse, they are actually all relatively genetically similar. So how do we define genetic diversity, and how do we conserve it? (Image Credit: Emir Kaan Okutan, Pexels Licence, Image Cropped)
Biodiversity has become an immensely popular buzzword over the last few decades. Yet the concept of genetic diversity has been less present in everyday ecological conversations. So today I want to go through why genetic diversity is important, how we define it, and why there is often controversy about its application in conservation science.
Maintaining genetic diversity in a species’ population is often key to ensuring its survival. As a population decreases in size, whether due to habitat loss, climate change or other human drivers, maintaining high genetic diversity gets more difficult. A smaller population size means the probability that close relatives will mate and pass similar genetic variance to their offspring increases. Things get more troublesome when deleterious mutations pop up in small populations. Sometimes these mutations may cause increases in disease, or reduce an individual’s ability to reproduce. And even when they don’t have immediate effects, they could accumulate along generations just due to chance if no new individuals enter the population.
This has practical applications for conservation science. We would like to have a wide range of individuals within a species, to ensure they are generally healthier and likely to persist for longer. A diverse population has a better chance of being able to adapt to human pressures. High variance in alleles within their immune cells could also help them combat various types of pathogens, as opposed to individuals with identical genetic makeup, which could be easily wiped out by a pandemic as they have no ability to adapt.
Conservation practitioners are hence striving to have focal populations retaining as much genetic variation as possible. But how diverse should a population within a species be? How many nucleotides or genes should differ between one another in order to maintain a species capable of adaptation?
Measuring Genetic Diversity
The genomic diversity of all living species is enormous. Every species has a different genome size and every individual within that species has different variations of that genome. For every single human individual, there are supposedly around 3 million nucleotide sites in which people differ with one another. For a comparison, this article is roughly 5000 characters, and on a nucleotide = letter bases it doesn’t even hit 0.2% of the section capable of variation.
However, though the number of human individuals populating the planet is currently approaching 8 billion, our genetic diversity is much lower than our closest primate relatives; chimpanzees and gorillas. Genetic diversity is no guarantor of success, and there are many species that do just fine without it. Invasive species for instance, often thrive in new environments from quite a small initial population.
Jonathan Romiguier’s 2014 study showed that the amount of genetic variation is more determined by our life history traits than our population size. Going on this, it makes sense that the amount of genetic diversity required for a healthy population is quite species-specific (or perhaps life-history-specific). Mammals with high parental investment, low generational time, and small offspring size tend to have lower genetic diversity than bacteria that divides each minute.
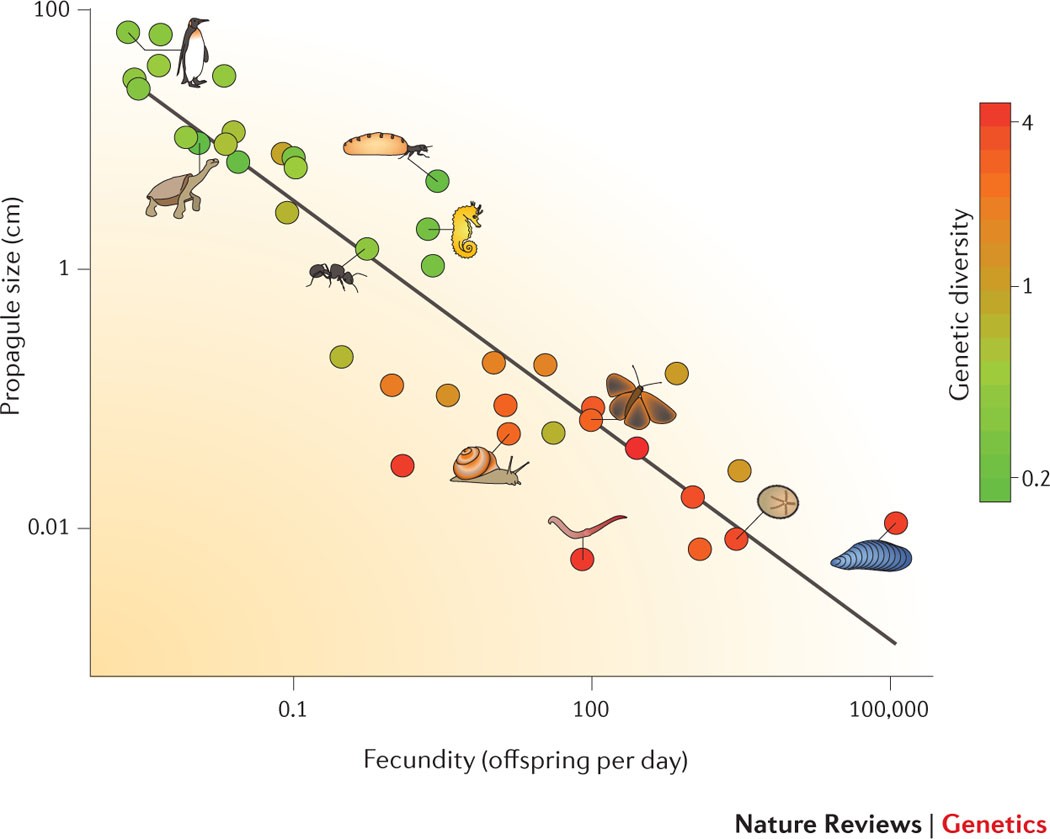
In this figure, you can see that as a species tends towards having less offspring and larger body sizes, they have lower genetic diversity. Source: Ellegren and Galtier (2016)
Genetic variation also does not necessarily correlate with diversity in their appearance. The African cichlid fish are an incredibly diverse array of species, and quite easy to tell apart, yet they are relatively genetically similar. A recent study also found that the various beak types of Darwin’s famous finches also difficult to distinguish at the genetic level. Also, have you heard about mouse lemurs? 24 different species, all living on Madagascar, and all really quite hard to tell apart.
As a result, when we discuss genetic diversity, we need to consider which part of the genome are we talking about. Of those billions of nucleotides of the species genome, different sections experience different kinds of processes. Some parts are easily mutated, some aren’t. Some parts move around, some don’t. There is also mitochondrial DNA that could only be inherited from the females. It’s a really complicated mess, and our understanding of it is still only just scratching the surface.

Whilst the 24 different species of mouse lemur all look quite similar, they are genetically quite varied (Image Credit: Bernard Dupont, CC BY-SA 2.0)
There are several ways to measure genetic diversity, and each way is capable of telling a different story. For example, if we sample areas that are close to genes which are responsible for important processes and as a result more important to a species’ ability to adapt, we will generally obtain lower diversity than in areas unaffected by natural selection. There’s no harm in changing a few bases that don’t affect our ability to digest glucose, for example. And the decisions we’ve had to make in estimating genetic diversity only get more complicated the more we learn about the evolutionary history of a species.
That last point is really the kicker: though we’re constantly making genome sequencing cheaper and obtaining higher quantities of genetic data, it unfortunately doesn’t make measuring genetic diversity for conservation purposes any easier. Different kinds of genetic markers provide different information, and not everyone measures genetic diversity the same way. A popular solution is to assess all aspects of the genome, but this is dependent on research budget and questions. A recent research on Darwin’s finches found that before their extinctions, some populations still retained relatively high genetic variation; genetic diversity does not necessarily save species.
Scientists also have mixed opinions on using genome-wide data to measure genetic diversity: some argue that it is overkill and bandwagon science, some say it is the only way to get the full picture. In any case, whilst our ability to map genomes charges forward, I hope our understanding of the results can match it.
Sabhrina Gita Aninta is a conservation geneticist currently pursuing her PhD at Queen Mary University of London to understand how genome-wide variation of the endemic pigs and buffalos from Southeast Asia could assist their conservation. Follow her Twitter here for an update of her work, along with a mix of conservation, biodiversity, evolution, but mostly various rants and random stuffs in Indonesian and English.

Pingback: How you can help ancient ungulate conservation using ancient DNA | Ecology for the Masses
Pingback: The Numbers Game of Species Conservation | Ecology for the Masses